2. Xingcheng Sanatorium of Shenyang Military Region
Radiation-induced thymic lymphoma (RITL) is one of the classic models in radiation carcinogenesis[1, 2, 3, 4]. Exposure to ionizing radiation caused cancer initiation and promoted cancer progression[5, 6, 7]. Oncogene Ras and tumor suppressor gene p53 played important roles in radiation-induced carcinogenesis[5]. The previous study showed that ERK1/2,STAT3 and SHP-2 were involved in γ-ray radiation-induced thymic lymphomas in BALB/c mice[8].
MicroRNAs (miRNAs) is a class of small RNAs approximately 18-22 nucleotides in length. MiRNAs suppressed protein expression by inhibiting translation or inducing mRNA degradation by binding to the 3'-untranslational region (3'UTR) of target mRNAs[9]. Deregulation or dysfunction of miRNAs contributes to cancer development[9, 10, 11, 12, 13, 14]. Our previous studies proved that miR-21 played an important role in RITL by directly targeting tumor suppressor gene Big-h3[15],and down regulation of miR-200c promoted RITL by targeting BMI1[16].
MiR-203 is identified as a stemness inhibiting miRNA that is highly expressed in the epidermis where it targets α and β isoforms of TP63 to promote epidermis differentiation[17, 18]. MiR-203 has also been shown to be aberrantly expressed in several types of human cancers including bladder,colon,pancreatic,liver,prostate and lung tumors[19, 20, 21, 22, 23].
Importantly,miR-203 is repressed by transcriptional repressor zinc-finger E-box binding homeobox1 (ZEB1),a repressor of multiple key mediators of epithelial differentiation[24],and a potent activator of epithelial-mesenchymal transition (EMT)[25]. EMT is a key program during cancer development and has been linked to tumor invasion,metastasis and chemo-resistance[26]. However,the role of miR-203 in RITL has not been fully investigated.
In this study,the role of miR-203 in RITL was investigated by up or down regulated the level of miR-203,then cell proliferation and apoptosis were evaluated. In the last part,the potential targeted genes are predicated and confirmed,and hoped to provide a new mechanism for radiation-induced thymic lymphoma.
BALB/c mice were obtained from Second Military Medical University,China. All mice were housed in a specific pathogen free facility for all experiments. All animal experiments were undertaken in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals"[27] with the approval of the Laboratory Animal Center of the Second Military Medical University,Shanghai. All efforts were made to reduce the number and suffering of animals.
2.Total-body irradiationA 60Co irradiator was used for total-body ionizing irradiation. Un-anaesthetized mice were placed in well-ventilated plastic boxes and exposed to the γ-ray irradiation at a distance of 3 m from the source. Sub-lethal doses of 1.75 Gy γ-ray irradiation were delivered at a dose rate of 0.58 Gy/min as described previously[28, 29, 30, 31]. The mice were then removed from the plastic box and allowed free access to food and water. On average,70% of all mice died within 12-35 weeks after the last irradiation (from all causes),but the most important cause of death was thymic lymphomas,whereas the non-irradiated control mice all lived to 35 weeks later.
3.Cell culture and transfectionNIH3T3,EL4 and HEK293 were purchased from Cell Bank of Chinese Academy of Science (Shanghai,China). These cell lines were maintained in Dulbecco's modified Eagle's medium (Invitrogen,USA) in the presence of 10% heat-inactivated fetal bovine serum,100 U/ml penicillin and 100 μg/ml streptomycin in a humidified 5% (V/V) atmosphere of CO2 at 37℃. Transfection was performed using Lipofectamine 2000 reagent (Invitrogen,USA) according to the manufacturer's protocol. Cells were seeded in 24-well at 6×104 cells per well and transfected with miRNA mimics or miRNA antisense oligonucleotides(ASO) at a concentration of 50 nmol/L.
4.RNA extraction and qRT-PCRRNA from thymic tissues of different groups were homogenized in TRIzol (Invitrogen,USA) and isolated according to the manufacturer's instruction. Reverse transcription and real-time PCR were subsequently performed in triplicate using the miScriptRT Kit and miScript PCR system (Qiagen,Germany) as described previously,while GAPDH was used as an internal control[32]. The primers were provided by Qiagen Company. Also,the miR-203 level was detected by using the Roche technology methods[16].
5.Methyl thiazolyl tetrazolium(MTT) assayFor MTT assay,500 cells per well were seeded in triplicate in a 96-well plate with complete growth medium. Cells were counted over 5 d using the MTT assay (Promega,USA) as described previously[12, 33, 34]. The data were measured by Microtiter plate reader 570 nm filters (Promega,USA).
6.Apoptosis assayCells were labeled with Annexin V-FITC and propidium iodide using an apoptosis detecting kit (Invitrogen,USA)following the manufacturer's instructions. Samples were analyzed by fluorescence activated cell sorter(FACS) assays (Becton Dickinson,USA)[35].
7.MiRNAs target predictionTargetScanHuman (http://www.targetscan.org/vert_61/)[36, 37, 38, 39] was applied to identify the potential target of miR-203.
8.MiRNAs mimics,miRNAs antisense oligonucleotides and overexpression plasmidsMiRNAs mimics and miRNAs ASO were obtained from GenePharma (China). MiRNAs mimics,negative control (NC) were transfected into cells at a concentration of 50 nmol/L using Lipofectamine 2000 transfection reagent according to the manufacturer's instructions. 48 h later,cells were collected for further experiments. The overexpression plasmid,pcDNA3.1-TBK1 (TANK-binding kinase 1),pcDNA3.1-SNAI2SLUG,pcDNA3.1-CCND1(cyclin D1) and pcDNA3.1-NRAS,were constructed and confirmed by Shenggong Company (Shanghai,China).
9. 3'UTR reporter analysisThe TBK1,EMT,regulatory factor SNAI2 and CCND1 3'UTR reporter plasmids (pRL-TBK1,pRL-SNAI2 and pRLCCND1) were constructed by Shenggong Company (Shanghai,China). Mutation in the miR-203 seed regions of the 3'UTR were generated using QuikChang multi-site-directed mutagenesis kit (Promega,USA). RL reporter plasmids (3.6 fmol) and pGL3-control (500 ng for normalization) were transfected with Lipofectamine 2000 into NIH3T3 cells (6×104 cells per well). Cells were collected after 48 h for assay using the Dual Luciferase reporter assay system (Promega,USA)[40].
10.Statistical analysisData were presented as the x±s form at least three independent experiments. The difference between two groups was analyzed using two-tailed Student's t-test. The difference between groups was analyzed using ANOVA analysis when more than three groups were compared. Correlation analysis was performed by two-tailed Pearson's correlation coefficient analysis. Statistical analyses were performed using SPSS software (Version 17.0). P<0.05 was considered significantly different.
To investigate the role of miR-203,the expression level of miR-203 was assayed firstly in RITL samples. Microarray analysis indicated that the miR-203 level in RITL tissues was lower than that in matched adjacent normal tissues (Figure 1). The mean expression level of miR-203 in RITL was lower than that in normal control. These data suggested that miR-203 might play roles in RITL.
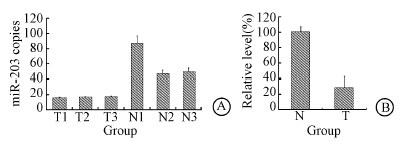 | Figure 1 Low expression level of miR-203 in RITL tissues,the miR-203 level in RITL (A) and the relative level of miR-203 in RITL (B) |
The miR-203 level was up-regulated in NIH3T3 and EL4 cell lines. The results showed 24 h after the miR-203 mimics transfection,the miR-203 level in NIH3T3 and EL4 cell lines were up-regulated (Figure 2). MiR-203 level was down-regulated in NIH3T3 and EL4 cell lines. 24 h after miR-203 ASO transfection,the miR-203 level in both cell lines were down-regulated. Cell apoptosis was assayed by FACS. Data showed that overexpression of miR-203 induced cells apoptosis and vice versa.Then cells proliferation was assayed by MTT,which showed that miR-203 mimic's transfection strongly inhibited the proliferation of both NIH3T3 cells and EL4 cells (Figure 3). Cell proliferation was promoted by miR-203 ASO transfection.
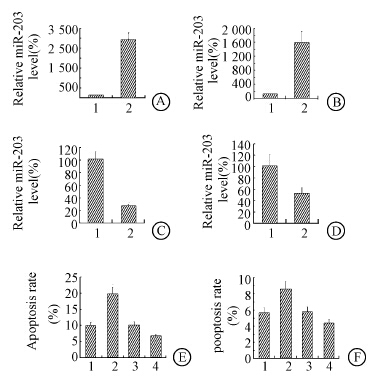 | Note:1.miR-NC; 2.miR-203; 3:miR-NC ASO; 4:miR-203 ASOFigure 2 MiR-203 mimics transfection inhibited cells proliferation and induced apoptosis and vice versa MiR-203 mimics transfection up-regulated the miR-203 expression in NIH3T3(A) and EL4(B); MiR-203 ASO transfection down-regulated the miR-203 expression in NIH3T3(C) and EL4(D); Up-regulation of miR-203 induced cells apoptosis and vice versa in NIH3T3(E) and EL4(F) |
 | Figure 3 MiR-203 targeted 3' UTR of TBK1,SNAI2 and CCND1 MiR-203 mimics transfection promoted NIH3T3(A) and EL4(B) cells growth; MiR-203 ASO transfection inhibited NIH3T3(C) and EL4(D) cells growth |
To investigate the involved mechanism of miR-203 in pathogenesis of RITL,potential target genes of miR-203 were predicated by bioinformatics algorithm. TBK1,SNAI2 and CCND1 were chosen for further study,and the 3'UTR of TBK1,SNAI2 and CCND1 were mutated (Figure 4).Then the 3'UTR of the three genes were cloned into luciferase reporter plasmids. MiR-203 and the reporter plasmids were co-transfected into HEK293 cells. The data showed that the 3'UTR of TBK1,SNAI2 and CCND1 were targeted by miR-203 (Figure 5). Then,TBK1,SNAI2 and CCND1 protein levels were assayed by Western blot after miR-203 transfection. TBK1,SNAI2 and CCND1 protein levels were down-regulated by miR-203 transfection. Thus data above indicated that TBK1,SNAI2 and CCND1 were targeted by miR-203.
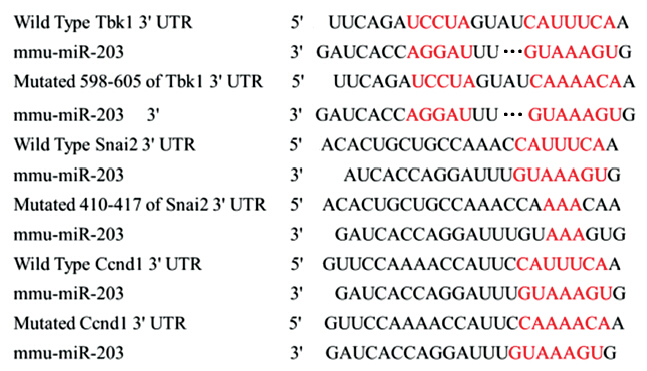 | Figure 4 The binding site of putative targeted gene,and mutated site of TBK1,SNAI2 and CCND1 |
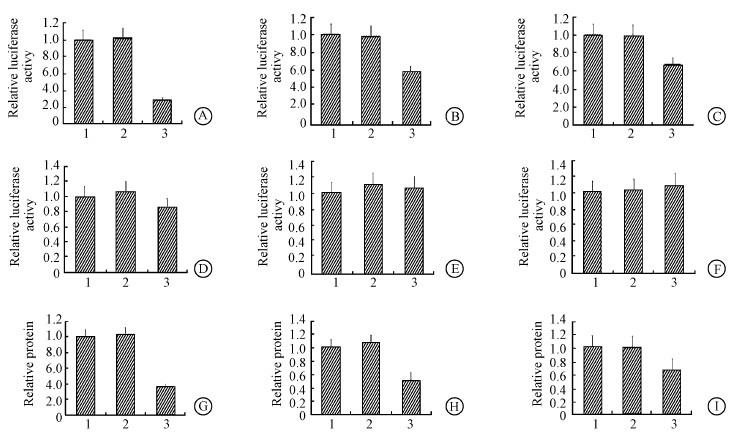 | Note:1.No miRNA; 2.miR-NC; 3.miR-203Figure 5 3'UTR reporter analysis of TBK1(A),SNAI2(B) and CCND1(C); 3'UTR reporter analysis of the mutated TBK1(D),SNAI2(E) and CCND1(F); MiR-203 mimics transfection inhibited TBK1(G),SNAI2(H) and CCND1(I) protein level |
TBK1,SNAI2/CCND1 and miR-203 mimics were co-transfected into NIH3T3 cell lines. 24 h later cell viability and apoptosis were assayed by trypan blue exclusion test and FACS analysis. Overexpression of TBK1,SNAI2 and CCND1 all partly showed rescue effects in cell viability and apoptosis,and the effect of miR-203 was partly restored by TBK1,SNAI2 and CCND1 overexpression plasmid separately (Figure 6). Then levels of TBK1,SNAI2 and CCND1 mRNA in 10 RITL were assayed. TBK1,SNAI2 and CCND1 mRNA level were higher than those in adjacent normal tissues. Next miR-203 level was examined in the 10 RITL. The miR-203 level in adjacent normal control tissues was arbitrarily defined as 100%. The correlation between miR-203 and the three genes were revealed by Pearson's correlation coefficient analysis: TBK1,SNAI2 and CCND1 mRNA levels were inversely correlated with miR-203 level in RITL (Figure 6). Thus the data indicated that miR-203 play a role in RITL via TBK1,SNAI2 and CCND1.
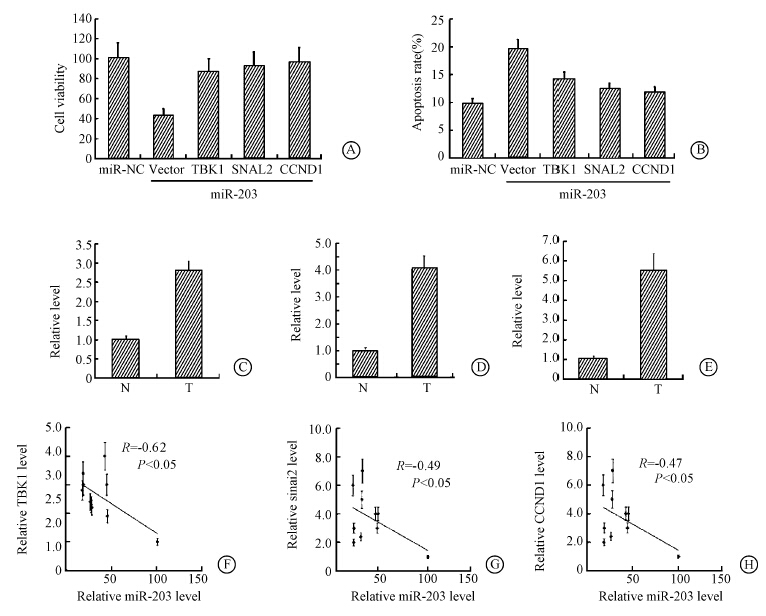 | Figure 6 Rescue effect of TBK1,SNAI2 and CCND1 Overexpression of TBK1,SNAI2 and CCND1 all partly showed rescue effects in cell viability(A) and apoptosis(B); TBK1(C),SNAI2(D) and CCND1(E) mRNA level were higher than adjacent normal tissues; TBK1(F),SNAI2(G) and CCND1(H) mRNA level were inversely correlated with miR-203 level in RITL |
Many miRNAs take part in the radiation-induced thymic lymphoma,such as miR-21[15] and miR-200c[16]. To our best knowledge,miR-203 played an important role in RITL.
MiR-203 has been reported as a tumor suppressor in basal cell carcinoma[41]. The research proved that down regulation of miR-203 promoted RITL indicating and functioned as a tumor suppressor in RITL.
In this study,overexpression of miR-203 proved to inhibit strongly NIH3T3 cells and EL4 cells proliferation and vice versa,which was also consistent with previous study[41],overexpression of miR-203 reduced proliferation and caused a delay in G1-to S phase transition. Whether miR-203 has an impact on cell cycle in RITL need further investigation. In the two studies above,the targeted genes of miR-203 were different. The data showed that miR-203 played a role in RITL via TBK1,SNAI2 and CCND1. The other studies showed that miR-203 targeted c-JUN. These data indicated that under different microenvironment,different genes may be targeted by miR-203[41]. The underlying mechanism is needed to further study.
Another study proved that obesity could increase IKKε and TBK1 activity in liver through NF-κB,and in turn initiated a program of counter-inflammation that preserved energy storage[42],which revealing the anti-inflammation role of TBK1. In RITL,the expression of TBK1 was higher than that in adjacent normal control tissues. As there was a close connection between inflammation and tumorigenesis,anti-inflammation role of TBK1 in RITL needs further study.
MiR-203 proved to down-regulate in RITL tissues,and overexpression of miR-203 strongly inhibited the cell proliferation cells via TBK1,SNAI2 and CCND1. The research provides a new mechanism for radiation-induced thymic lymphoma worth further investigation.
| [1] | Kaplan HS. Experimental alteration of host response to an occult tumor virus: induction of thymic lymphosarcomas in X-irradiated mice[J]. Natl Cancer Inst Monogr, 1960, 4: 141-150. |
| [2] | Kaplan HS. The role of radiation on experimental leukemogenesis[J]. Natl Cancer Inst Monogr, 1964, 14: 207-220. |
| [3] | Finn OJ, Boniver J, Kaplan HS. Induction, establishment in vitro, and characterization of functional, antigen-specific, carrier-primed murine T-cell lymphomas[J]. Proc Natl Acad Sci USA, 1979, 76 (8): 4033-4037. |
| [4] | Herranz M, Martín-Caballero J, Fraga MF, et al. The novel DNA methylation inhibitor zebularine is effective against the development of murine T-cell lymphoma[J]. Blood, 2006, 107 (3): 1174-1177. |
| [5] | Little JB. Radiation carcinogenesis[J]. Carcinogenesis, 2000, 21 (3): 397-404. |
| [6] | Potworowski EF, Gagnon F, Beauchemin C, et al. Dendritic cells prevent radiation-induced thymic lymphoma[J]. Leukemia, 1996, 10 (10): 1639-1647. |
| [7] | Shin SC, Kang YM, Kim HS. Life span and thymic lymphoma incidence in high- and low-dose-rate irradiated AKR/J mice and commonly expressed genes[J]. Radiat Res, 2010, 174 (3): 341-346. |
| [8] | Fu Z, Huang D, Cai J, et al. Expression changes of ERK1/2, STAT3 and SHP-2 in bone marrow cells from γ-ray induced leukemia mice[J]. J Radiat Res, 2006, 47 (2): 121-130. |
| [9] | Ambros V. The functions of animal microRNAs[J]. Nature, 2004, 431 (7006): 350-355. |
| [10] | Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer[J]. Annu Rev Med, 2009, 60: 167-179. |
| [11] | Hou J, Lin L, Zhou W, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma[J]. Cancer Cell, 2011, 19 (2): 232-243. |
| [12] | Li D, Liu X, Lin L, et al. MicroRNA-99a inhibits hepatocellular carcinoma growth and correlates with prognosis of patients with hepatocellular carcinoma[J]. J Biol Chem, 2011, 286 (42): 36677-36685. |
| [13] | Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease[J]. Cell, 2012, 148 (6): 1172-1187. |
| [14] | Croce CM. Causes and consequences of microRNA dysregulation in cancer[J]. Nat Rev Genet, 2009, 10 (10): 704-714. |
| [15] | Liu C, Li B, Cheng Y, et al. MiR-21 plays an important role in radiation induced carcinogenesis in BALB/c mice by directly targeting the tumor suppressor gene Big-h3[J]. Int J Biol Sci, 2011, 7 (3): 347-363. |
| [16] | Cui J, Cheng Y, Zhang P, et al. Down regulation of miR200c promotes radiation-induced thymic lymphoma by targeting BMI1[J]. J Cell Biochem, 2014, 115 (6): 1033-1042. |
| [17] | Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, et al. MiR-203 represses ‘stemness’ by repressing ΔNp63[J]. Cell Death Differ, 2008, 15 (7): 1187-1195. |
| [18] | Yi R, Poy MN, Stoffel M, et al. A skin microRNA promotes differentiation by repressing ‘stemness’[J]. Nature, 2008, 452 (7184): 225-229. |
| [19] | Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer[J]. Cancer Res, 2007, 67 (18): 8699-8707. |
| [20] | Greither T, Grochola LF, Udelnow A,et al. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival[J]. Int J Cancer, 2010, 126 (1): 73-80. |
| [21] | Furuta M, Kozaki KI, Tanaka S, et al. MiR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma[J]. Carcinogenesis, 2010, 31 (5): 766-776. |
| [22] | Garofalo M, Romano G, Di Leva G, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers[J]. Nat Med, 2012, 18 (1): 74-82. |
| [23] | DeCastro AJ, Dunphy KA, Hutchinson J, et al. MiR203 mediates subversion of stem cell properties during mammary epithelial differentiation via repression of ΔNP63α and promotes mesenchymal-to-epithelial transition[J]. Cell Death Dis, 2013, 4: e514. |
| [24] | Aigner K, Dampier B, Descovich L, et al. The transcription factor ZEB1 (ΔEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity[J]. Oncogene, 2007, 26 (49): 6979-6988. |
| [25] | Burk U, Schubert J, Wellner U, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells[J]. EMBO Rep, 2008, 9 (6): 582-589. |
| [26] | Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease[J]. Cell, 2009, 139 (5): 871-890. |
| [27] | Editors National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals.Guide for the care and use of laboratory animals[S]. Source 8th edition. Washington: National Academies Press, 2011. |
| [28] | Fu Z, Huang D, Cai J, et al. Expression changes of ERK1/2, STAT3 and SHP-2 in bone marrow cells from γ-ray induced leukemia mice[J]. J Radiat Res (Tokyo), 2006, 47 (2): 121-130. |
| [29] | Liu C, Gao F, Li B, et al. TLR4 knockout protects mice from radiation-induced thymic lymphoma by down regulation of IL6 and miR-21[J]. Leukemia, 2011, 25 (9): 1516-1519. |
| [30] | Liu C, Zhang C, Mitchel RE, et al. A critical role of toll-like receptor 4 (TLR4) and its' in vivo ligands in basal radio-resistance[J]. Cell Death Dis, 2013, 4: e649. |
| [31] | Li B, Zhang C, He F, et al. GSK-3β inhibition attenuates LPS-induced death but aggravates radiation-induced death via down-regulation of IL-6[J]. Cell Physiol Biochem, 2013, 32 (6): 1720-1728. |
| [32] | Wu N, Zhang C, Bai C, et al. MiR-4782-3p inhibited non-small cell lung cancer growth via USP14[J]. Cell Physiol Biochem, 2014, 33 (2): 457-467. |
| [33] | van Meerloo J, Kaspers GJ, Cloos J. Cell sensitivity assays: the MTT assay[J]. Methods Mol Biol, 2011, 731: 237-245. |
| [34] | Han ZB, Yang Z, Chi Y, et al. MicroRNA-124 suppresses breast cancer cell growth and motility by targeting CD151[J]. Cell Physiol Biochem, 2013, 31 (6): 823-832. |
| [35] | Lu J, Wen M, Huang Y, et al. C2ORF40 suppresses breast cancer cell proliferation and invasion through modulating expression of M phase cell cycle genes[J]. Epigenetics, 2013, 8 (6): 571-583. |
| [36] | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets[J]. Cell, 2005, 120 (1): 15-20. |
| [37] | Friedman RC, Farh KK, Burge CB, et al. Most mammalian mRNAs are conserved targets of microRNAs[J]. Genome Res, 2009, 19 (1): 92-105. |
| [38] | Grimson A, Farh KK, Johnston WK, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing[J]. Mol Cell, 2007, 27 (1): 91-105. |
| [39] | Garcia DM, Baek D, Shin C, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs[J]. Nat Struct Mol Biol, 2011, 18 (10): 1139-1146. |
| [40] | Grentzmann G, Ingram JA, Kelly PJ, et al. A dual-luciferase reporter system for studying recoding signals[J]. RNA, 1998, 4 (4): 479-486. |
| [41] | Sonkoly E, Lovén J, Xu N, et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma[J]. Oncogenesis, 2012, 1: e3. |
| [42] | Reilly SM, Chiang SH, Decker SJ, et al. An inhibitor of the protein kinases TBK1 and IKK-varepsilon improves obesity-related metabolic dysfunctions in mice[J]. Nat Med, 2013, 19 (3): 313-321. |
 2015, Vol. 35
2015, Vol. 35


